
I am a Juniorprofessor at Georg-August University Göttingen and a member of the Institute of Computer Science and CIDAS. I lead the research group for Computational Cell Analytics since March 2022. My research is focused on deep learning methods for computer vision, in particular segmentation, applied primarily to microscopy images.
Previously, I have worked as a PostDoc at EMBL Heidelberg in the group of Anna Kreshuk and did my PhD at the University of Heidelberg under the supervision of Anna Kreshuk and Fred Hamprecht. During my PhD and PostDoc I have mainly worked on instance segmentation problems, with a focus on large volumetric electron microscopy data.
constantin.pape@informatik.uni-goettingen.de
Institute of Computer Science
Georg-August University Göttingen
Jobs, thesis, student projects
I am currenrly not offering any master thesis, bachelor thesis or internship projects, but will offer new projects for the summer term 2024. You can check out previous projects here: here.
Research
Our group's research goals is to develop deep learning based methods for advanced bioimage analysis. The goals include developing segmentation and tracking methods that require minimal supervision, interactive segmentation and tracking, as well as self-supervised representation learning for bioimage data. We are further working on methods for protein structure reconstruction in identification from Cryo-EM and expansion microscopy data, and develop approaches for building shared represetations of imaging, genetic and clinical data.
The adoption of deep learning has improved the quality of image analysis for microscopy dramatically in the last decade. In the same period, the throughput, field of view and time resolution in microscopy has increaded significantly, requiring the automation of key analysis steps, such as the identification of cells through segmentation, following cells over time through tracking or classifying cells into distinct states. While deep-learning based methods yield high enough quality to achieve automation for many applications they require large amounts of (manually) annotated training data . This limitation makes them impractical for applications where no such training data is available and is too costly to create. Fortunately, the last years have seen reseach interest in weak supervision, domain adaptation and self-supervision, which promises to make training of high-quality models with significantly less annotated data possible. However, these methods are mostly developed for natural images and their adoption in microscopy has so far been limited. We aim to bridge this gap, building on initial work for domain adaptation, weak supervision and even reinforcement learning.
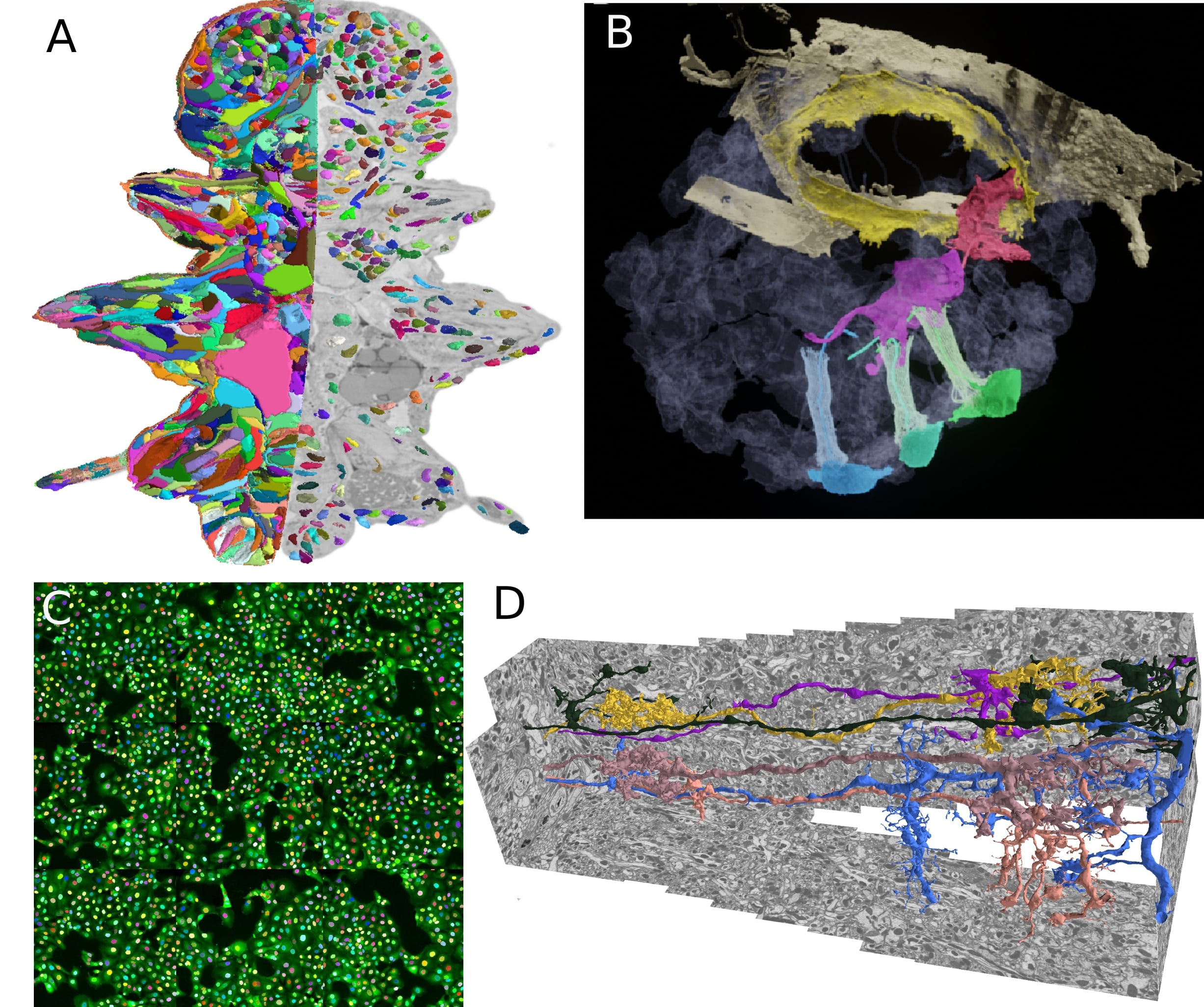
My previous research has been focused on boundary based instance segmentation. I have developed graph-based methods based on globally optimal graph partitioning and fast heuristic partitioning. The initial focus of my work has been neuron segmentation in electron micrscopy (D), for which I have developed methods that scale to multi terayte volumes and can incorporate biological priors. These methods have been used in various life science applications: building a high resolution genetic and morphological atlas of P. dumerilii (A), analyzing the morphology of precursor neural cells in sponges (B) or developing an imaging based SARS-CoV-2 antibody assay (C). I plan to continue this work and make these methods available through easy-to-use tools in order to democratize the access to large-scale volumetric segmentation.
I am dedicated to open source and open science and am actively contributing to several related efforts. In particular, the bioimage.io modelzoo, a resource to share deep learning models for microscopy image analysis and ome.ngff, a new image data format that supports efficient storage of large data and on-demand access in the cloud. Furthermore, I co-develop MoBIE, a Fiji plugin for exploring and sharing large multi-modal image data.
Foundation models for bioimaging
Foundation models have become a focus of deep learning research in the last years. The term describes large architectures that were trained on large diverse datasets and can then be applied to solve many different tasks in a given field. These models have first become available in natural language processing (e.g. GPT3 and GPT4), but are now also emerging in computer vision (e.g.
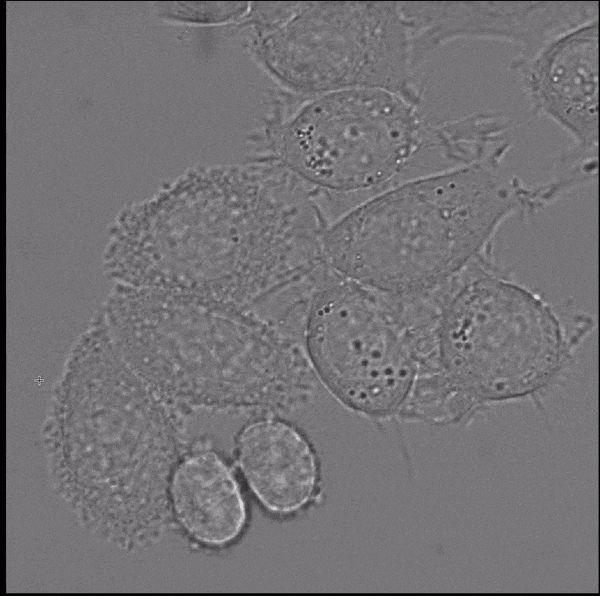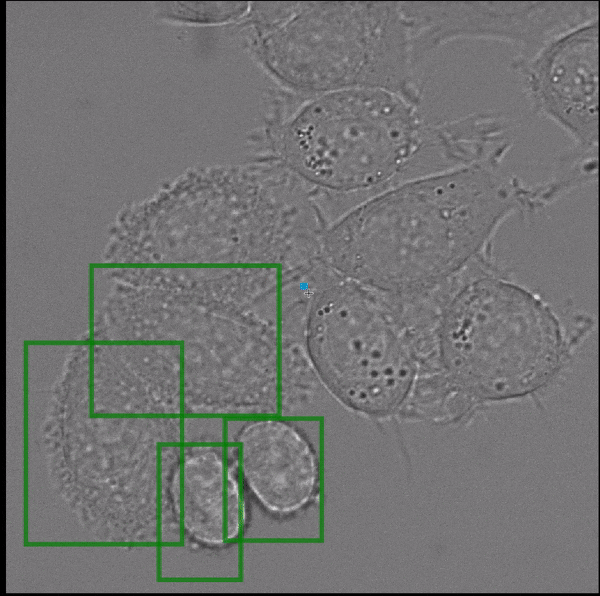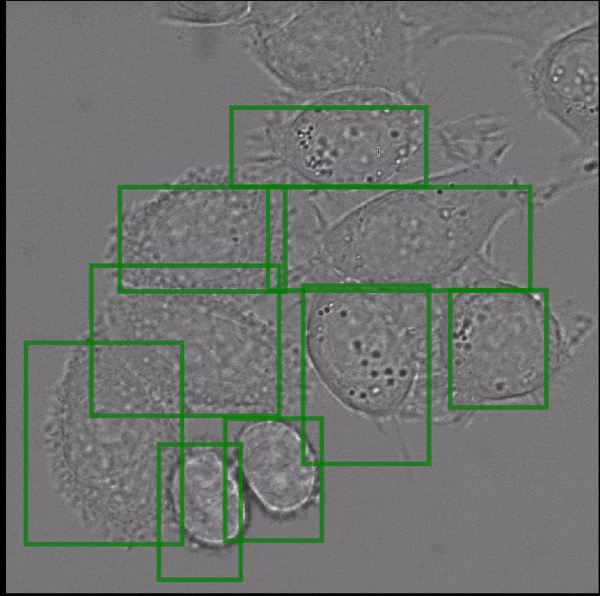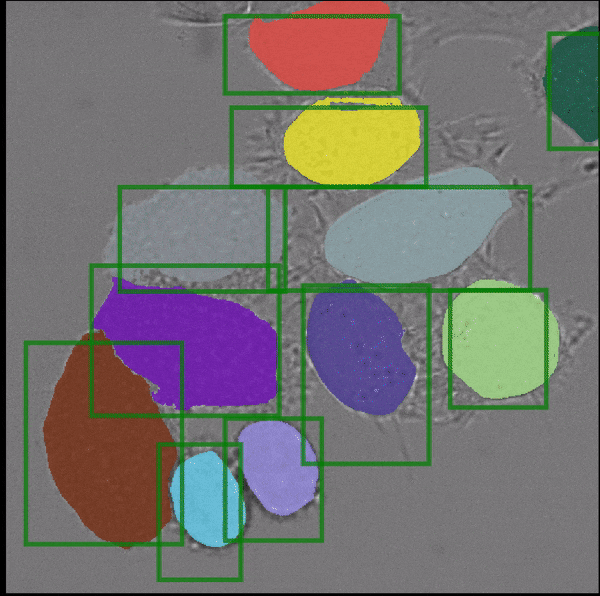
Segment Anything, a recently published vision foundation model, enables fully automated and interactive instance segmentation and works for a wide set of image modalities. We have found that it works well for microscopy data, even without further fine-tuning, and can speed up data annotation significantly. Motivated by this finding we are now developing Segment Anything for Microscopy, a data annotation tool for biologists. The animation shows how it is ued to interactively segment cells from bounding box annotations.
The goal of this project is to further improve foundation models like Segment Anything for bioimaging data and develop them into an universal solution for segmentation and tracking problems. To achieve this goal you will:
- Develop methods to fine-tune Segment Anything (or similar models) on large microscopy datasets to improve their performance for our target applications. Building on recent methods for fine-tuning language foundation models such as LoRA.
- Implement few-shot segmentation approaches that enable learning from interactive segmentation results and using them for automatic segmentation (and tracking) without having to train a new model.
- Build and improve tools for interactive segmentation and tracking based on (fine-tuned) vision foundation models.
This project is funded by the Multiscale Bioimaging Cluster of Excellence (MBExC). The methods will be developed in collaboration with other research groups within MBExC and will be applied to solve challenging image analysis problems they are facing.
Teaching
I hold a lecture on deep learning and offer seminars on applications of deep learning at the University of Göttingen. See the next paragraph for a short overview of these courses and check out UniVZ for the courses currently offered.
In addition to teaching at the university I co-organize the Deep Learning for Image Analysis course at EMBL, see the last course page for details. I am also a Hertha Sponer College Instructor, where I teach a course on advanced image analysis (course details will follow soon).
Deep Learning for Computer Vision
The lecture covers deep learning and its applications to computer vision. The following topics are covered:
- Fundamentals of deep learning (artificial neural networks, training via stochastic gradient descent)
- Convolutional neural networks and their application to image recongnition
- Deep learning approaches for vision tasks: segmentation, object detection, denoising, etc.
- Generative models (Variational Autoencoders, Generative Adverserial Networks) and their applications in image synthesis
- Semi-supervised, weakly-supervised and self-supervised learning for image data
- Vision transfomers
- Deep learning for videos, point clouds and scene rendering
Deep Learning in Biology and Medicine
The seminar discusses advanced topics in applications of deep learning methods in biology and medicine. We cover applications in image analysis, structural biology (e.g. protein folding with Alpha Fold), large language models in medicine and more. The seminar is offered every summer term.
Publications
Please find the full list of publications on Google Scholar. Below is a list of selected publications:From our group:
- Segment Anything for Microscopy, bioRxiv (2023), Archit, ..., Pape
- Probabilistic Domain Adaptation for Biomedical Image Segmentation, arXiv (2023), Archit and Pape
From my PhD and PostDoc:
- MoBIE: a Fiji plugin for sharing and exploration of multi-modal cloud-hosted big image data, Nature Methods (2023), Pape, Meechan et al.
- From Shallow to Deep: Exploiting Feature-Based Classifiers for Domain Adaptation in Semantic Segmentation, Frontiers in Computer Science (2022), Matskevych, Wolny, Pape and Kreshuk
- Profiling cellular diversity in sponges informs animal cell type and nervous system evolution, Science (2021), Musser, Schippers, Mizzon, Kohn, Pape et al.
- Whole-body integration of gene expression and single-cell morphology, Cell (2021), Vergara, Pape, Meechan et al.
- Microscopy-based assay for semi-quantitative detection of SARS-CoV-2 specific antibdoes in human sera, BioEssays (2021), Pape, Remme et al.
- Leveraging domain Knowledge to improve microscopy image segmentation with lifted multicuts, Frontiers in Computer Science (2019), Pape et al.
- The mutex watershed: efficient, paramter-free image partitioing, ECCV (2018), Wolf, Pape et al.
- Solving large multicut problems for connectomics via domain decomposition, ICCV Workshops (2017), Pape et al.
- Multicut brings automated neurite segmentation closer to human performance, Nature Methods (2017), Beier, Pape et al.
CV
- 2010-2013 Bachelor of Science, Physics, Ruprecht-Karls University Heidelberg
- 2013-2016 Master of Science, Physics, Ruprecht-Karls University Heidelberg
- 2016-2021 PhD, Physics, Ruprecht-Karls University Heidelberg
- 2017-2018 Visiting scientist, Janelia Research Campus
- 2018-2021 Visiting scientist, EMBL Heidelberg
- 2021-2022 PostDoctoral Fellow, EMBL Heidelberg
- 2022- Juniorprofessor, Georg-August University Goettingen